GENETIC HETEROGENEITY /
TUMOR EVOLUTION
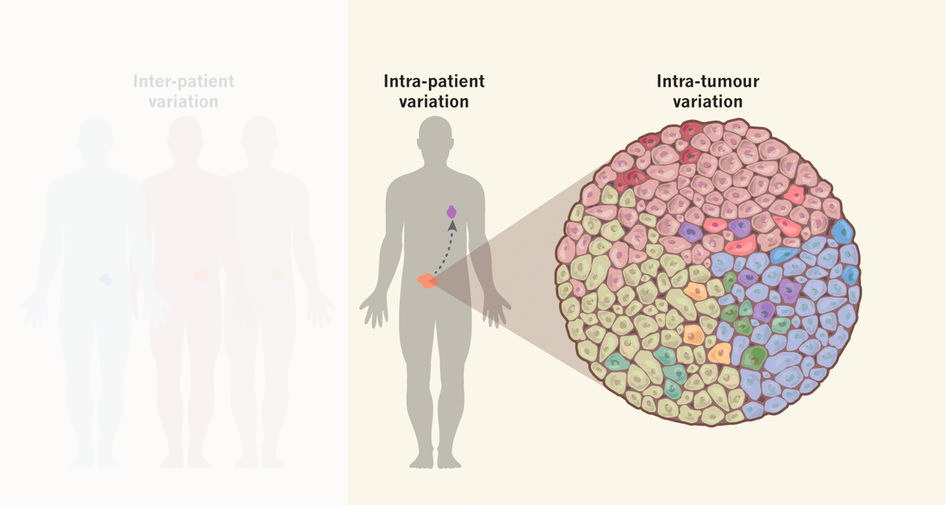
The vast majority of tumors is genetically heterogeneous, meaning that cancers are composed of many different genetic subclones. In recent years, continuous innovation in sequencing techniques has been the main driver to advance our knowledge about genetic heterogeneity in cancers. Unfortunately, little is known about tumor evolution at cell cycle resolution.
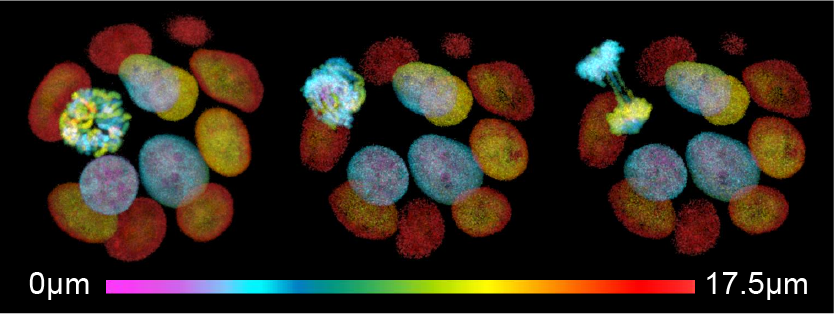
Organoids are currently the closest representatives of primary human tumors that are compatible with high temporal resolution imaging of cellular events. Our lab has established real-time imaging of patient-derived tumor organoids over periods of days to monitor their evolutionary process – from a single cell to an established tumor population – and examine the level of chromosomal instability in human cancer organoids, as well as the tumor specific tolerance levels to mitotic errors.
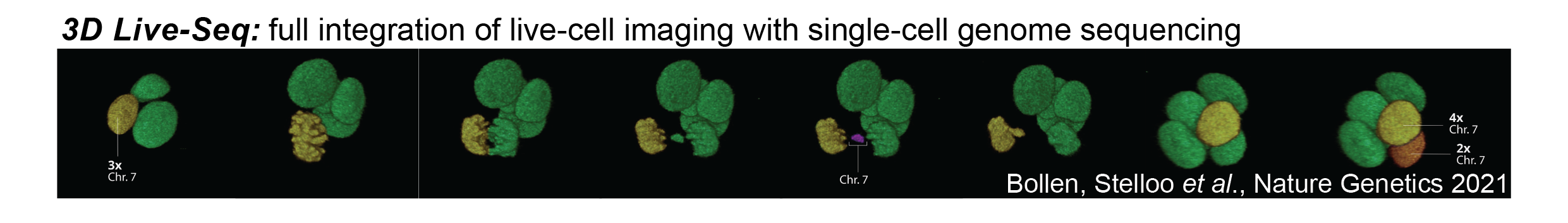
Although the research fields of cell biology and genetics are well-established, there is relatively little interaction between both disciplines. Most notably, we recently managed to directly integrate live-cell microscopy of 3D organoids with single cell DNA sequencing of those filmed cells to address immediate genetic consequences of aberrant cellular processes, like errors during mitosis. We dubbed this technology 3D LiveSeq. Most extreme, 3D LiveSeq enables reconstruction of the tempo and patterns by which copy-number alterations emerge and propagate in tumor tissue across multiple consecutive cell cycles as if we are monitoring cancer genome evolution in real-time.
We now aim to utilize 3D LiveSeq to study genome evolution during malignant transformation, as well as to continue its innovation to map emergence and propagation of cancer mutations at base-pair resolution.
- Drost et al., Nature 2015
- Bolhaqueiro et al., Nat Genet 2019
- Bollen, Stelloo et al., Nat Genet 2021