UNDERSTANDING (SINGLE-CELL) DRUG RESPONSE USING PATIENT-DERIVED TUMOR ORGANOIDS
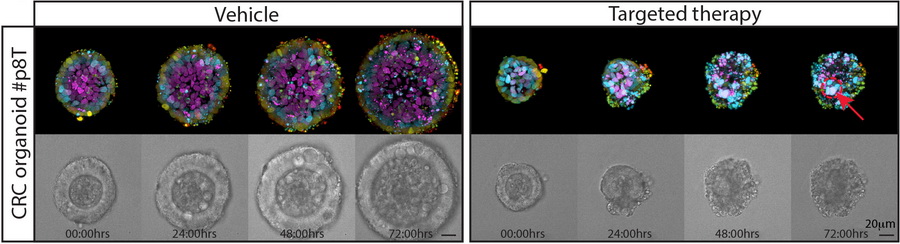
Regularly, anti-cancer therapies are effective against the majority of tumor cells. Unfortunately, there is frequently a small population of cells that shows resistance against the applied therapy. Little is known about the nature and origin of these resistant cells.
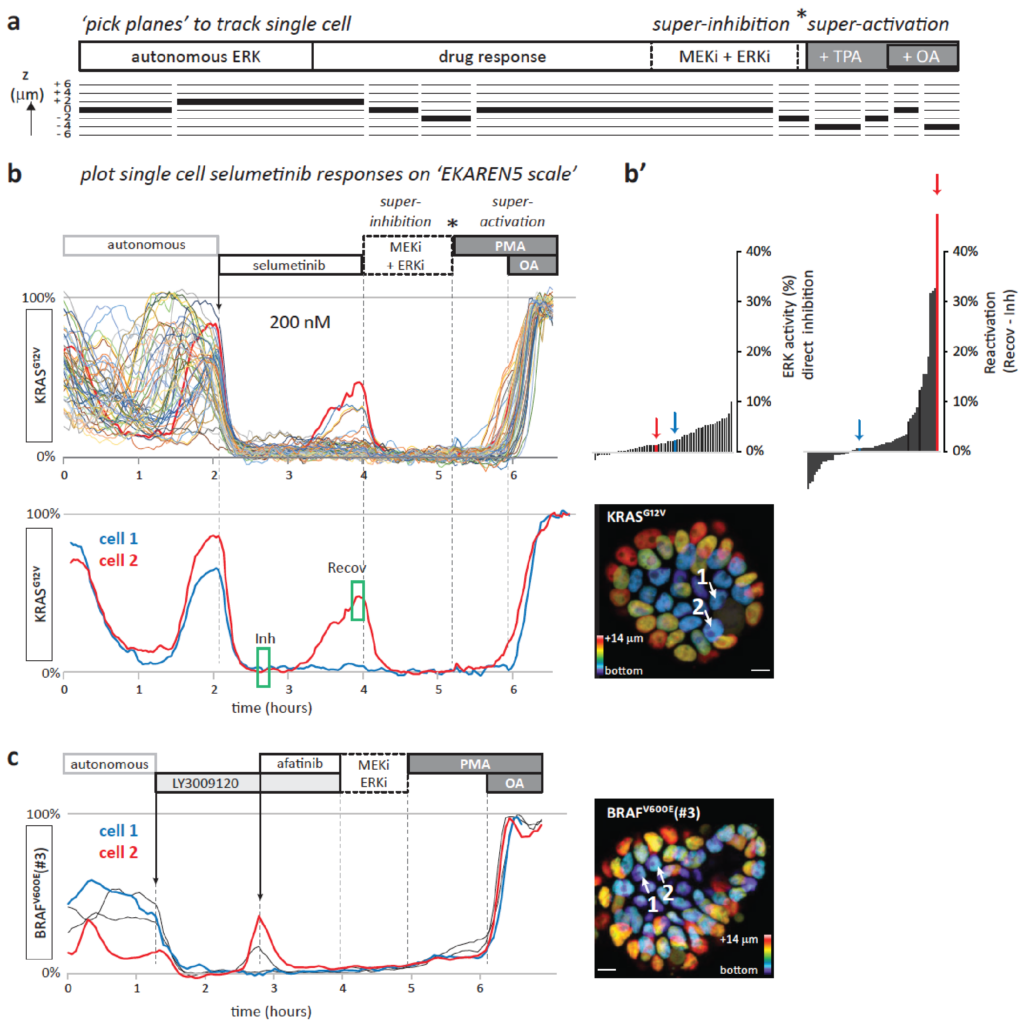
Functional heterogeneity within the population of tumor cells underlies the wide variety of cellular responses against therapies. Patient-derived tumor organoids recapitulate the histopathological features of native tumors, including patient specific responses towards therapies. Using established biobanks of patient-derived tumor organoid, we perform drug screens to understand genotype-phenotype correlations, in particular concerning RAS pathway mutations. In addition to classical high-throughput drug screens, we develop new strategies for imaging-based drug sensitivity measurements. Most recently, we developed real-time single-cell drug response measurements in patient-derived tumor organoids using FRET-based quantitative read-out of ERK activity, the downstream effector protein of the RAS pathway. We are intrigued by single-cell drug responses towards targeted inhibitors of the RAS pathway in the context of population dynamics and therapy resistance.
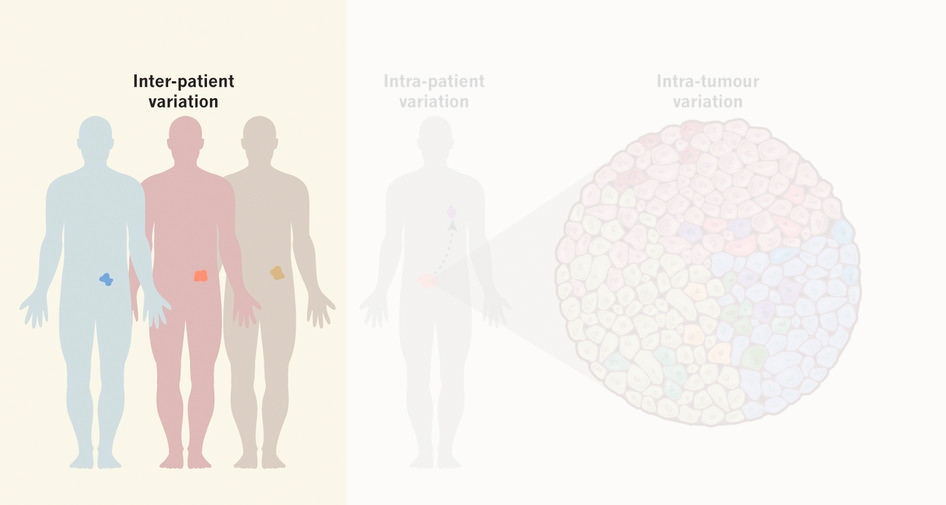
Despite similarity in ‘common’ cancer mutations, there is often variability in drug response between patients’ tumors (inter-patient heterogeneity). We apply molecular genetics to engineer cancer organoids to our own interest, which includes CRISPR/Cas9-mediated homologous recombination to introduce or correct cancer mutations, to study and cross-compare their influence on drug sensitivity.
- Verissimo, Overmeer, Ponsioen et al., eLIFE 2016
- Kopper et al., Nat Med 2019
- Ponsioen et al., Nature Cell Biology 2021
- Mertens et al., Cell reports 2023